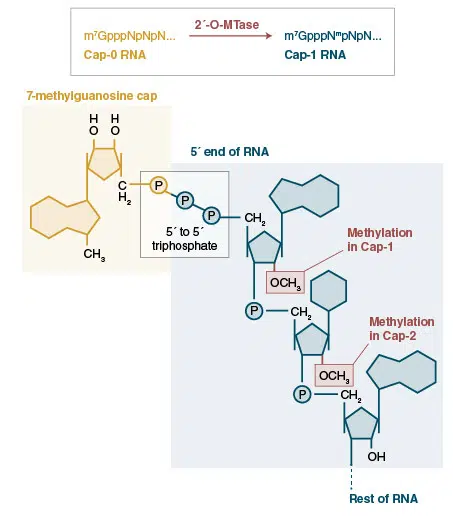
There are many examples in nature of modified RNAs, for instance, in eukaryotic organisms, mRNAs are characteristically modified by the post-transcriptional addition of an inverted 7-methyl-G to form a cap structure that is critical for the maintainence of mRNA stability and for ensuring engagement of the mRNA translation machinery. Another example of modification occurs on the 3′-ends of some classes of small regulatory RNAs (piwi interacting RNAs and endogenous small interfering RNAs in animals, and plant microRNAs) are modified by the addition of a methyl group to the 2′-position of the ribose on the terminal nucleotide. This modification is essential for piRNA stability and regulatory function.
Enzymes that modify RNA are important tools to characterize RNA species, because their activity can differ based upon existing modifications. For instance, 5′-capped RNAs are not substrates for polynucleotide kinases, or RNA ligases, but are substrates for decapping enzymes. Similarly, ligation and polyadenylation efficiency for 2′-O-methylated 3′-ends of RNAs differs from those with unmodified 3′-ends. The selective properties of RNA modifying enzymes enable researchers to distinguish these RNA species. Some unique end modifications can be used to selectively degrade or isolate particular RNA species when treated with the correct series of enzymes.
In vitro modification of RNA ends can be particularly important to prepare RNA for downstream applications such as adaptor ligations in preparation for sequencing platforms where certain end modifications are required for efficient representation (1).
Modification of RNA bases, e.g. 5 methyl cytosine, or N6 methyladenine, to evade innate immune responses in mRNA transfected cells is important in experiments that use this technique to influence cell fate.
Further information can be found in our Technical Resources section or at neb.com. Information on trademarks can be found here.