The NEBNext ARTIC kits meet the increasing need for reliable, accurate and fast methods for sequencing SARSCoV-2, with Oxford Nanopore Technologies or Illumina sequencing. The kits use the ARTIC multiplexed ampliconbased whole-viral-genome sequencing approach and were developed in collaboration with the ARTIC Network (artic. network). The kits include balanced V3 primer pools for improved uniformity of genome coverage and are effective with 10-10,000 SARS-CoV-2 genome copies. The reagents and protocols for RT-PCR and library prep are all optimized specifically for the SARS-CoV-2 ARTIC workflows.
Improved sequence coverage for current virus variants and express protocols.
Starting September 2021, two options for primers are now available, in the NEBNext ARTIC SARS-CoV-2 FS Library Prep Kit (Illumina) (NEB #E7658), NEBNext ARTIC SARS-CoV-2 Companion Kit (Oxford Nanopore Technologies) (NEB #E7660) and NEBNext ARTIC SARS-CoV-2 RT-PCR Module (NEB #E7626):
As of mid-February 2022, you will receive both the “ARTIC V3” primer pool and the VarSkip Short v2 primers (VSSv2) for free with NEBNext ARTIC reagents (not applicable to #E7650). Use the VSSv2 primers to achieve excellent coverage of circulating SARS-CoV-2 variants, including the Omicron and Delta variants. For robust coverage of the latest variants, we recommend using a spike-in primer pool in addition to the VSSv2 primers (see figure below). This pool is available separately on request.
Sequence coverage of SARS-CoV-2 genomes with VarSkip Short v2 plus spike-in primers
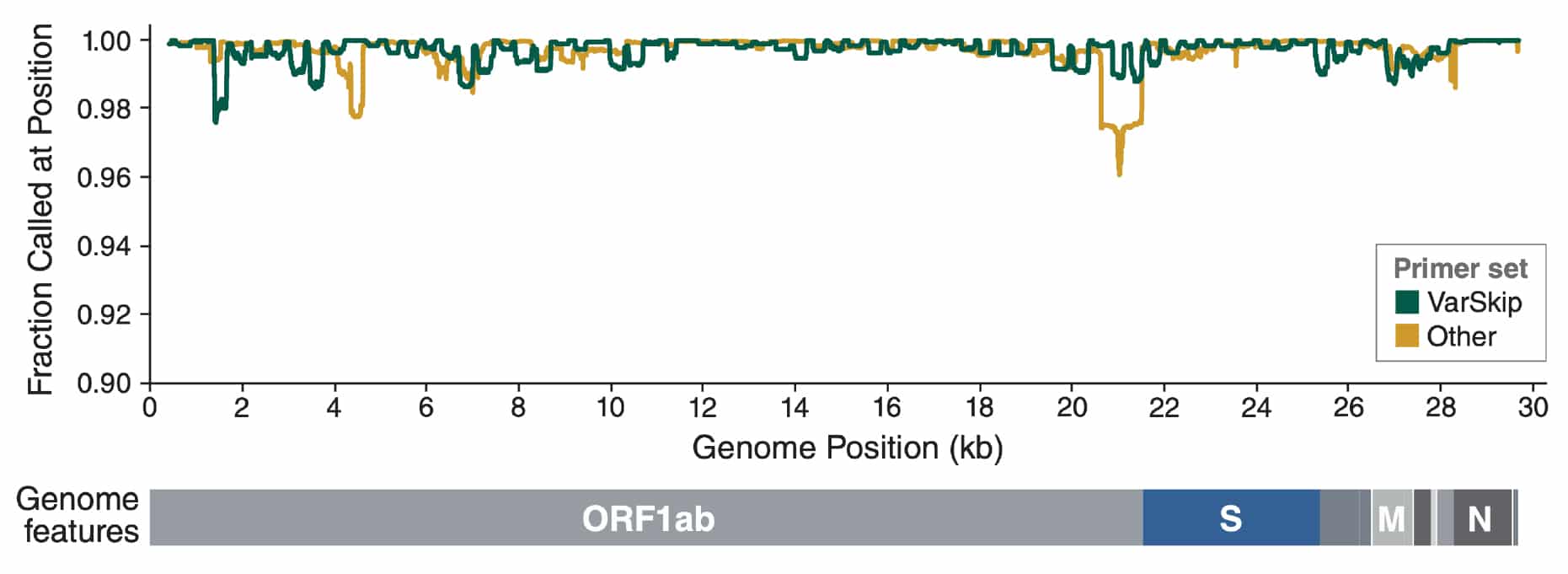
Consensus genomes known to be generated using VarSkip Short 2 (+ supplementary primers) (green) are compared with other genomes (yellow) submitted to Genbank between 2022-08-01 and 2022-09-24. Genomes were aligned to the NC_045512.2 reference (minimap2 -x map-ont -r 20000 –score-N=0, v.2.24). Ns were tabulated using Qualimap (v2.2.2).
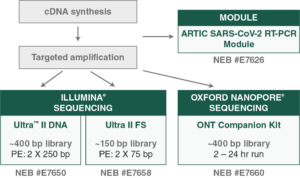
The two kits compatible with Illumina sequencing generate library inserts of ~150 bp (for 2 x 75 sequencing) or ~400 bp (for 2 x 250 sequencing), and include a novel DNA polymerase formulation that eliminates the need to normalize amplicon concentrations prior to library preparation.
- Improved uniformity of SARS-CoV-2 genome coverage
- Effective with a wide range of viral genome inputs (10–10,000 copies)
- Single RT conditions for all input amounts
- No amplicon normalization prior to library preparation (Illumina-compatible kits)
- NEBNext Sample Purification Beads (SPRIselect®) provided
- Cost-effective
NEBNext ARTIC Kit Options:
NEBNext ARTIC Kit Workflow:
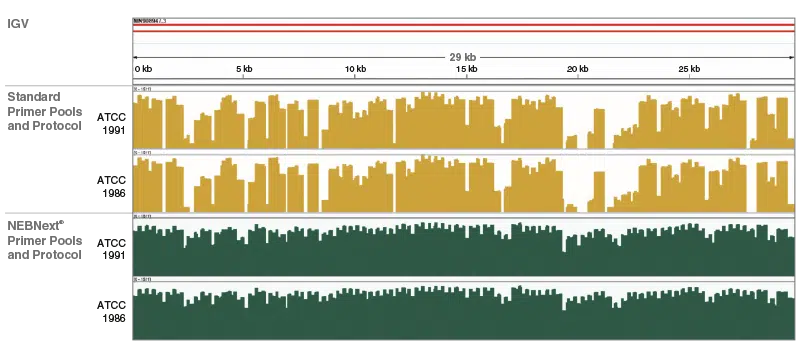
NEBNext ARTIC Kit Genome Coverage:
Integrative Genome Viewer visualization of read coverage across the SARS-CoV-2 genome show the superior genome coverage and uniformity using the NEBNext primers, reagents and protocols. For detailled experimental data, please visit www.neb.com/E7660
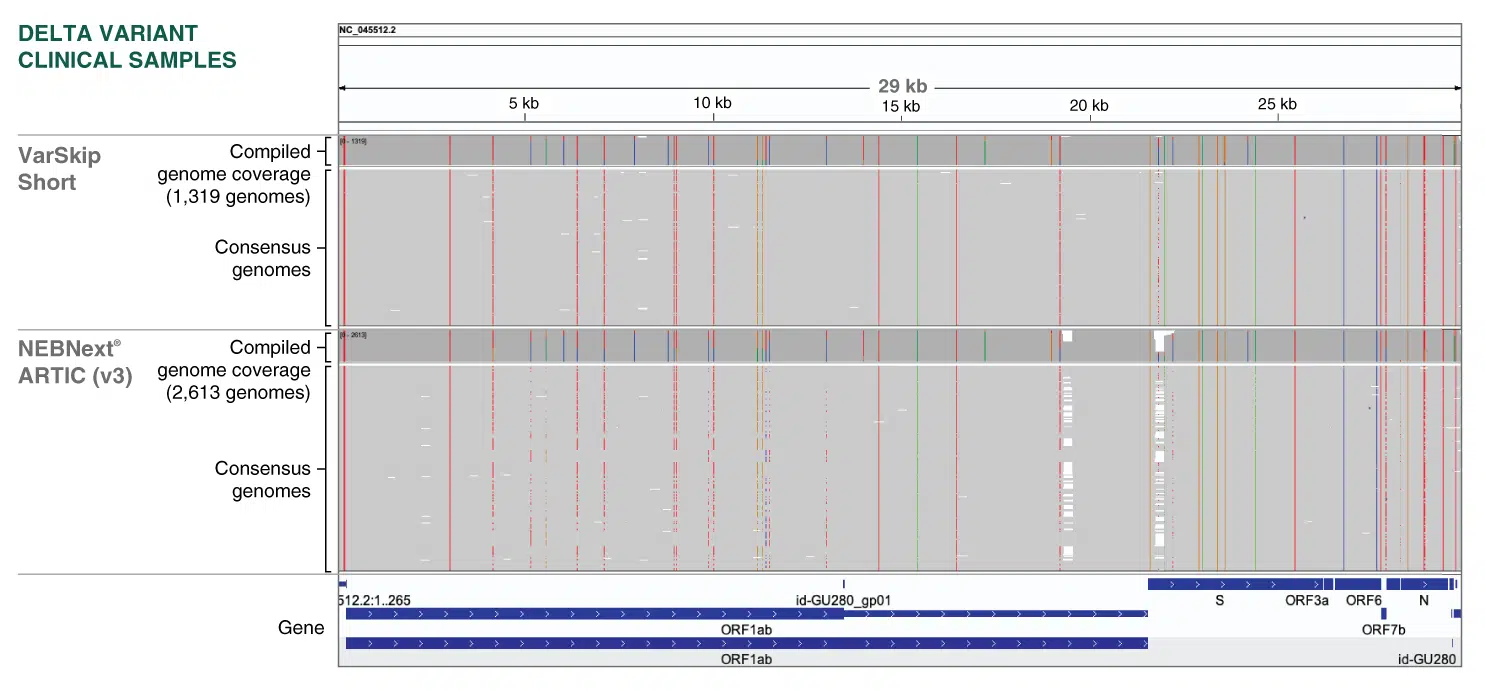
Improved sequence coverage of SARS-CoV-2 Delta clinical samples with VarSkip Short primers:
Consensus sequences classified as B.1.617.2 (Delta) in Massachusetts (where VarSkip Short was field tested) were collected from NBCBI Virus [1] and partitioned into 2,613 ARTIC V3 and 1,319 VarSkip Short sequences using the SRA “design” field (NEB_VarSkip_v1 = VarSkip Short, others assumed to be ARTIC V3). Reads were aligned to the NC_045512.2 reference using minimap2 (minimap2 -x asm5) and visualized using IGV. Consensus sequences generated from VarSkip amplicons suffered fewer dropouts. For detailled experimental data, please visit www.neb.com/E7658
|
„We have been impressed by the great performance of the NEBNext ARTIC FS kit in our sequencing of human samples containing SARS-CoV-2 RNA. With this kit we have been successfully processing samples of varying quality and quantity and have been amazed by its sensitivity and remarkable uniformity of the viral genome coverage. The quick turnaround time of the robust workflow has been truly beneficial for us.“ Dr. Vladimir Benes, Head of GeneCore EMBL Heidelberg, Early-Access-User |
||||
 |
||||
Product table
As of: 01.01.2024
Further information can be found in our Technical Resources section or at neb.com. Information on trademarks can be found here.