Reliably remove unwanted RNA species from your NGS samples
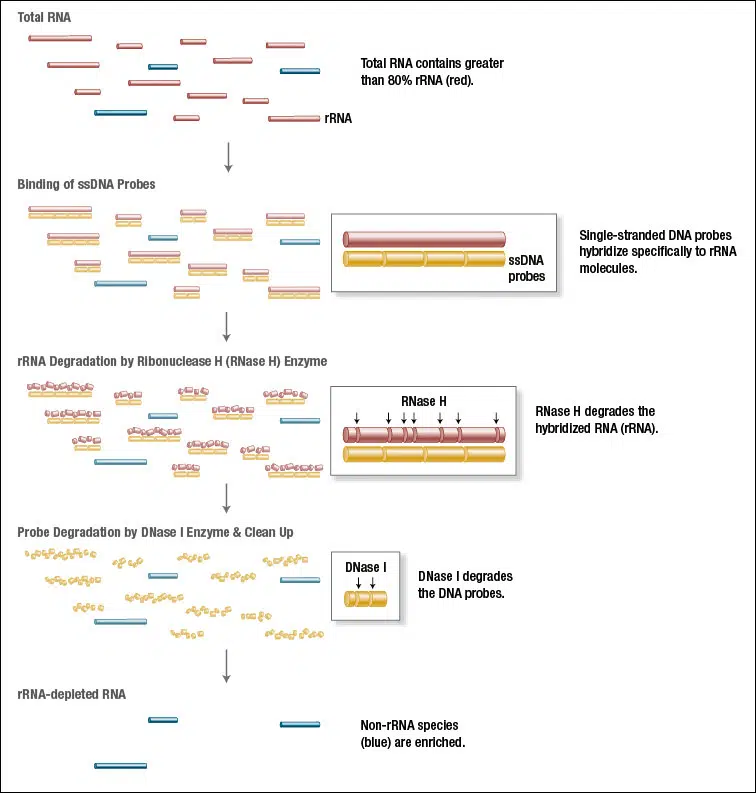
In typical RNA isolations the great majority of RNA comprises rRNA, while other RNAs like mRNAs are less abundant. Abundant RNAs can conceal the biological significance of less-abundant transcripts, making their efficient and specific removal desirable. NEBNext RNA Depletion kits facilitate the removal of abundant RNAs, while ensuring retention of RNAs of interest. These kits employ the efficient RNase H method: Single-stranded DNA probes hybridize specifically to rRNA molecules and subsequently RNase H degrades the hybridized RNA.
The special feature of the NEBNext kits: The DNA probes cover the entire rRNA sequence so that the probes can also hybridize to fragmented rRNA.
Therefore, our Next-Gen-Seq validated NEBNext rRNA Depeletion Kits can successfully and reliably be used even on poor quality total RNA samples (such as fragmented RNAs or RNA from FFPE material) starting from RIN >1!
Both non-coding regulatory RNA and fragmented mRNA are retained in the sample, enabling an unbiased glimpse of the transcriptome.
With the NEBNext rRNA Depletion Kits, you typically need standard input amounts of 10 ng to 1 µg of total RNA. The protocol takes only 2 hours of time with less than 10 min hands-on time.
NEBNext rRNA Depletion Kit Workflow
Please click to enlarge
- Suitable for degraded or high-quality (intact) RNA
- 10 ng to 1 μg total RNA input
- Remove over 98% of rRNA, and obtain more relevant sequence reads from your sample
- Obtain a more complete transcriptome picture through retention of noncoding and incomplete RNAs
- Easily integrated upstream of any downstream random-primed cDNA synthesis protocol
- Quick protocol: 2 hours, less than 10 min hands-on time
- Also available with RNA Sample Purification Beads
- Automation-friendly, already automated on various platforms
Application Note:
Selective depletion of abundant RNAs to enable transcriptome analysis of low-input and highly-degraded RNA from FFPE breast cancer samples
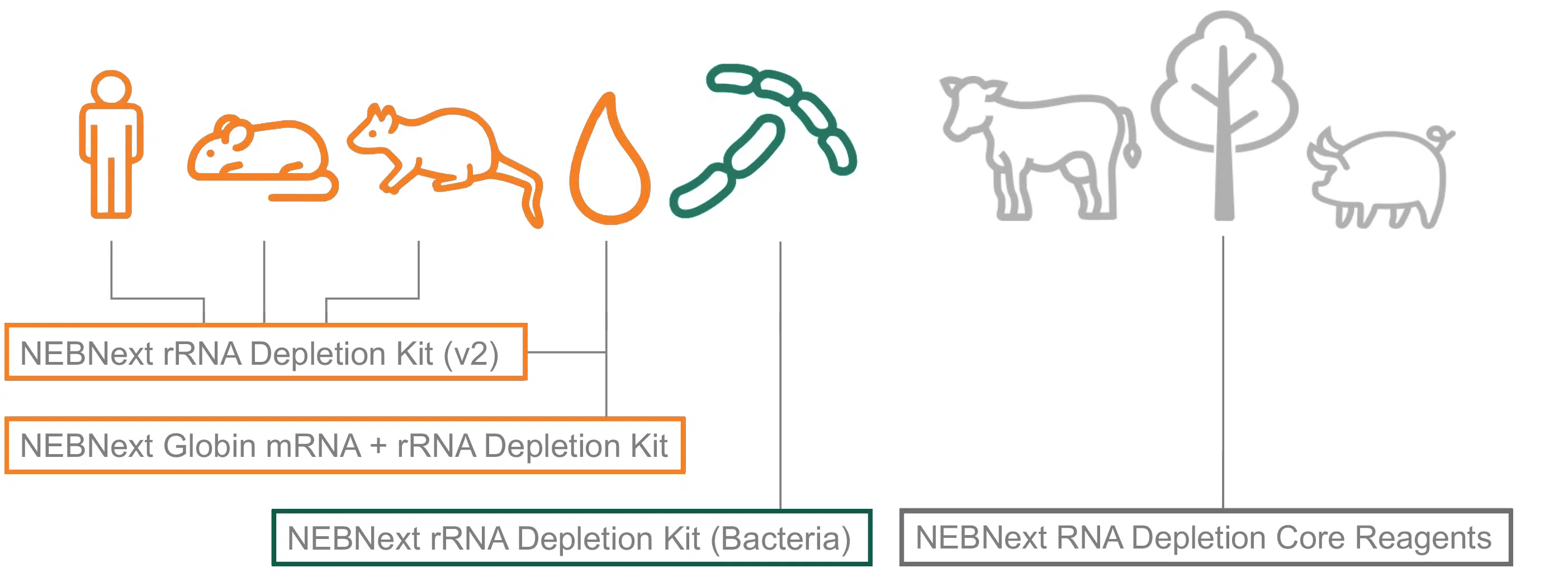
Choose the right Kit depending on your model organism/application
Our current NEBNext RNA Depletion products

NEBNext rRNA Depletion Kit (Human /mouse/rat) Version 2 (v2)
Suitable for use with total RNA preparations from human, mouse and rat samples, these kits are optimized for depletion of both cytoplasmic (5S, 5.8S, 18S, 28S, ITS and ETS) and mitochondrial (12S, 16S) ribosomal RNA, from intact and degraded samples.
- New in version 2: Also removes ITS and ETS fragments of the unwanted rRNA
- Perfect for samples from human, mouse and rat
- Suitable for degraded or high-quality (intact) RNA
- 10 ng to 1 μg total RNA input
- Quick protocol: 2 hours, less than 10 min hands-on time
Customer comments:
|
„… the NEB kit was the best performing kit that we have tested in our lab!“ Nikhil Bharti (AG Frau Prof. Ignatova), Biochemie, Uni Hamburg |
||||
 |
||||
The NEBNext rRNA Depletion Kit v2 enables the enrichment of desired RNA species from total human, mouse and rat total RNA.
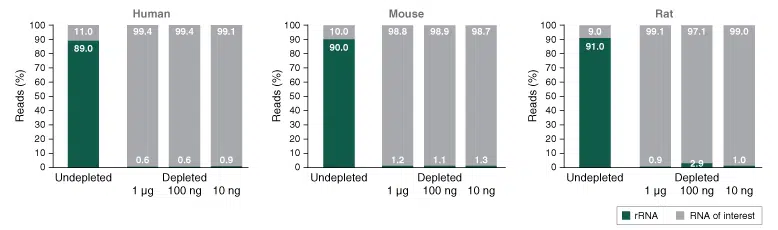
Efficient depletion of ribosomal RNAs from different input amounts (1 ug, 100 ng, 10 ng) of universal human, mouse and rat reference RNA. For subsequent library prep the NEBNext Ultra II Directional RNA Library Kit was used.

NEBNext Globin & rRNA Depletion Kit
In blood samples, the great majority of RNA comprises rRNA and globin mRNA, and their simultaneous removal is advantageous. The NEBNext Globin & rRNA Depletion Kit (Human/Mouse/Rat) depletes globin mRNA, cytoplasmic rRNA (5S, 5.8S, 18S, 28S, ITS and ETS) and mitochondrial rRNA (12S, 16S). The kit is effective with human, mouse and rat total RNA preparations, both intact and degraded.
- Compatible with total RNA from blood samples
- Specific and efficient depletion of globin mRNA and rRNA
- Can be combined with poly(A) mRNA isolation for additional removal of non-coding RNA
- Suitable for degraded or high-quality (intact) RNA
- 10 ng to 1 μg total RNA input
- Quick protocol: 2 hours, less than 10 min hands-on time
Efficient depletion of globin mRNA and ribosomal RNA from human, mouse and rat blood samples
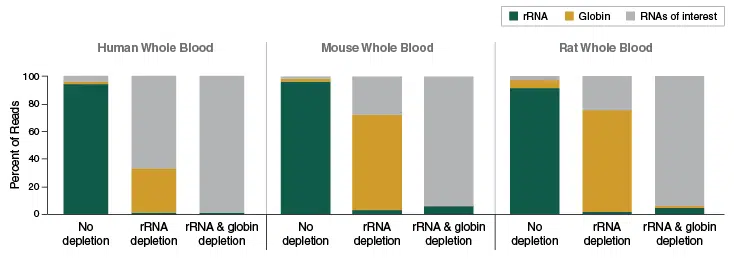
Depletion of globin mRNA and ribosomal RNA using the NEBNext Globin mRNA & rRNA Depletion Kit allows enrichment of desired RNA species from human, mouse and rat blood samples. For subsequent library prep the NEBNext Ultra II Directional RNA Library Kit was used.

NEBNext Bacterial rRNA Depletion Kit
Also bacterial RNA isolations are dominated by ribosomal RNA (rRNA). The NEBNext rRNA Depletion Kit (Bacteria) quickly and reliably removes unwanted rRNA (5S, 16S and 23S) from gram-positive and gram-negative bacterial samples. The kit can be used with monoculture or samples with mixed bacterial species (e.g. metatranscriptomics) with both intact or degraded RNA
Customer comments:
|
„The NEB Bacterial Depletion has depleted rRNA equally or better than our previous ribodepletion gold standard across a wide RIN quality range. We have been pleased with the flexibility of Total RNA input ranges and have routinely gotten effective ribodepletion at 100 ng Total RNA Input in both single isolates and metagenomic samples. The protocol is also more ergonomically friendly than bead based ribodepletion protocols. Of all the new bacterial ribodepletion methods we have tested, NEB was by far the best.“ Research Assistant, Biomedical research institution |
||||
 |
||||
- Specific and efficient depletion of bacterial rRNA
- Suitable for gram-positive and gram-negative species
- Compatible with monocultures or mixed bacterial species (“metatranscriptomics”)
- Suitable for degraded or high-quality (intact) RNA
- 10 ng to 1 μg total RNA input
- Quick protocol: 2 hours, less than 10 min hands-on time
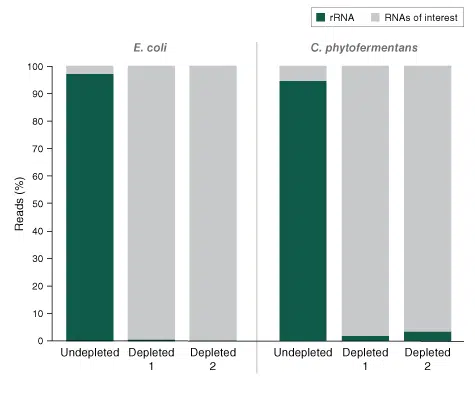
The NEBNext rRNA Depletion Kit for bacteria – one workflow for a range of different bacterial species
The NEBNext rRNA Depletion kit for bacteria is optimized for the depletion of ribosomal RNA from a range of different bacterial species, whether mono- or mixed cultures, gram-positive or gram-negative bacteria. Depletion of ribosomal RNA from 100 ng total RNA of Escherichia coli and Clostridum phtofermentans is shown as an example. The NEBNext Ultra II Directional RNA Library Prep Kit was used for subsequent library prep.

Custom RNA Depletion mit dem NEBNext RNA Depletion Core Reagent Set (available with or without purifications beads) – suitable for all species and RNAs!
Are you working with a sample type for which current RNA depletion kits are not compatible, or do you need to remove a specific RNA from your sample? NEBNext now enables a customizable approach to deplete unwanted RNA from any organism, using probe sequences designed with a user-friendly web tool.
- Customized RNA depletion from any organism using specific probes
- Easy design of probes with the NEBNext Custom RNA Depletion Design Tool
- Suitable for degraded or high-quality (intact) RNA
- 10 ng to 1 μg total RNA input
- Quick protocol: 2 hours, less than 10 min hands-on time
Step 1: Use the online NEBNext Custom RNA Depletion Design Tool to obtain custom probe sequences, by entering the sequence of your target RNA.
Step 2: Oder ssDNA probe oligonucleotides from your trusted oligo provider.
Step 3: Use the probes with the NEBNext Custom RNA Depletion Core Reagent Set or in combination with other NEBNext RNA Depletion Kits.
Customer comments:
|
„The open-access tool to design sequences for rRNA removal that NEB is providing to the research community has been very useful for us. This method for customization of RNA depletion has proven valuable for a range of organisms. Use of custom probes for Xenopus laevis ribosomal RNA worked very well, resulting in complete depletion. And supplementing the probes in the NEBNext rRNA Depletion Kit (Bacteria) with custom probes for the bacillus Eggerthella lenta led to 90% depletion of the ribosomal RNA.“ Dr. Vladimir Benes, Head of Genomics Core Facility, EMBL, Heidelberg |
||||
 |
||||
|
„This technology has been superb for eliminating the ribosomal RNA for a range of custom projects, including tracking novel viruses in mosquitoes, longitudinal profiling for astronauts, and host-pathogen interactions in COVID samples. Any species can now have a rich, in-depth profile created of its complex transcriptome, from non-coding RNAs to RNA modifications“ Dr. Christopher Mason, Weill Cornell Medicine, New York (USA) |
||||
 |
||||
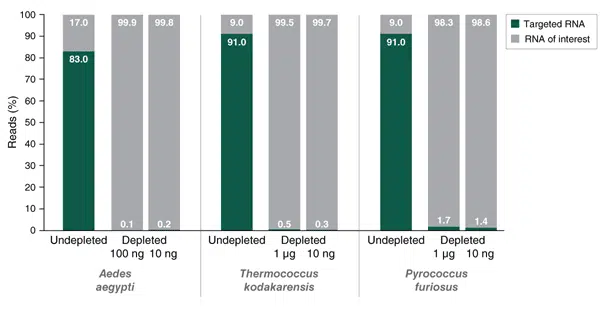
The NEBNext RNA Depletion Core Reagent Set enables a fully customized depletion of unwanted RNA species
The NEBNext Custom RNA Depletion Design Tool was used to design probes against rRNA of the mosquito Aedes aegypti, and the archaeal species Thermococcus kodakarensis and Pyrococcus furiosus. Total RNA (1 μg, 100 ng or 10 ng) was used as input for rRNA depletion using the Core RNA Depletion Reagent Set with the designed probes. The NEBNext Ultra II Directional RNA Library Prep Kit was used for subsequent library prep.
Erhältliche RNA Depletion Kits:
As of: 01.01.2023
NEBNext products are available at a permanent low price that can not be further discounted.
Further information can be found in our Technical Resources section or at neb.com. Information on trademarks can be found here.